Bitefirst
BiteFirst designs and builds bespoke highly secure web solutions involving complex forms and data management. Our client base includes university research departments, small businesses and charities.
We work with you from a blank sheet of paper to a secure, fully functioning solution delivering to tailored needs, whether that be complex large scale data analysis, workflow such as recruiting, auto email generation or bespoke reporting. We specialise in backend functionality enabling simple front end access, web design can be (and often is) included if a one stop solution is wanted. Based in both London and Norfolk, we can work in person, hybrid or fully remotely.
Led by a highly experienced technical expert with over 35 years of solution design and build and a Masters in cyber security, systems are built using current market toolsets, ensuring maintainability and security of both the data being used and any solution being built. Users are given secure access to data and depending on the solution, we can host the system and associated data, or it can reside within your technical estate.
Clinical Data
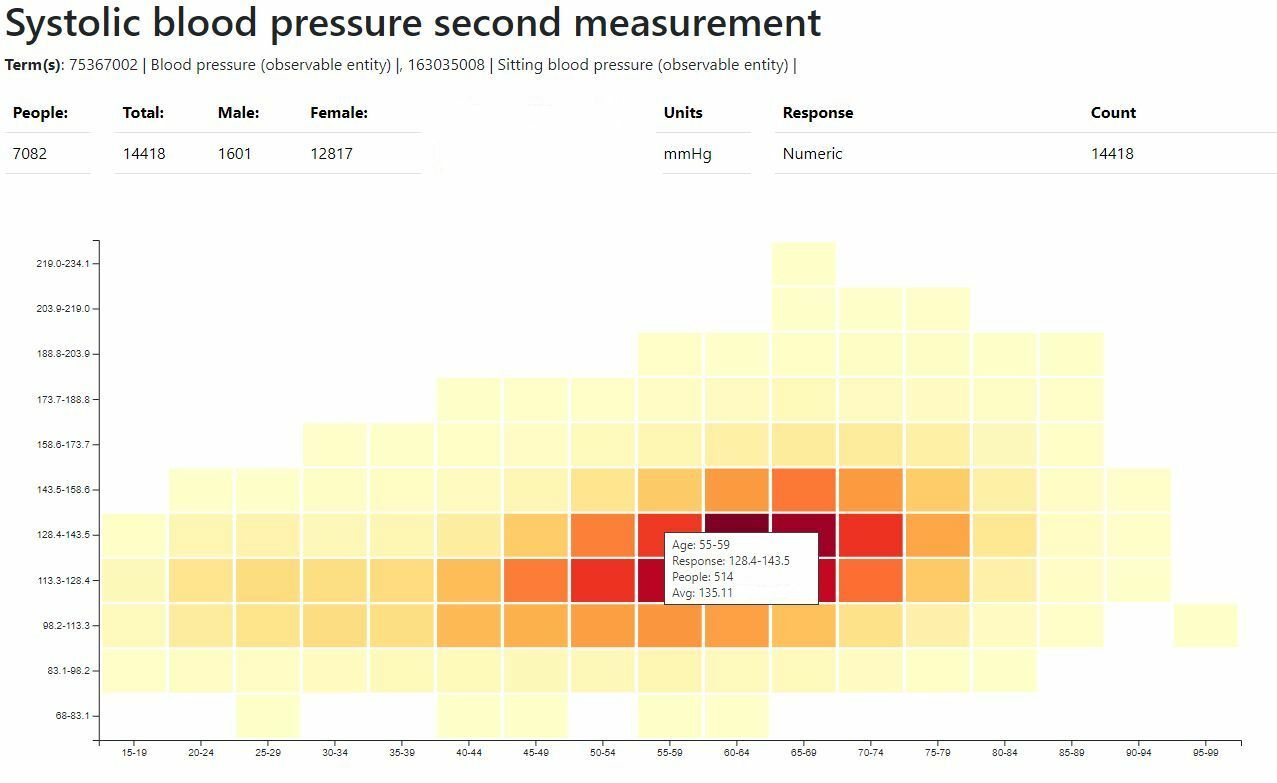
Bitefirst has completed systems for TwinksUK / Kings College London to support the onboarding of participants to medical studies, phenotype data management and research data requests. Standards such as the OMOP Common Data Model (CDM) and SNOMED thesaurus were adopted to aid future compatibility. See more detail here .
Bioinformatics
A request to fix a broken script at Imperial morphed into building a system to process tens of thousands of virus gene sequences using a machine learning tool to predict epitopes for use in vaccine evaluation and design. See more detail here .
Finance & Recruitment
Bitefirst built and maintains a recruitment system with both a public WEB site and a back-office solution to manage the recruitment process (client data, jobs, candidate data, adverts, payments etc). Other finance systems include an accounting system to support the company debt management process (data mining of billing systems, invoice production, workflow, letter production, court summons production, reporting etc.). See more detail here .
Technology
A broad set of technologies are used with a focus on an open source web stack of Bootstrap HTML with JavaScript or React JavaScript, a Ruby Rails API or Application server and a PostgreSQL database on an Ubuntu operating system. Python is often used for scripting, ETL, integration and gene sequence processing. Numerous tools and APIs are used (e.g. GitHub, image processing, NHS FHIR API, Google API, SharePoint API etc.).
Information Security
Systems are developed with skills gained from an Information Security MSc at Royal Holloway. This ranges from user authentication and authorisation through to firewalls, encryption and negating direct attacks such as SQL injection, cross site scripting and password attacks.
| Other Sections In This Site: |
|---|
| Sectors |
| Sectors / Clinical Data |
| Sectors / Bioinformatics |
| Sectors / Recruitment |
| Sectors / Billing |
| News |