Phenotype Storage
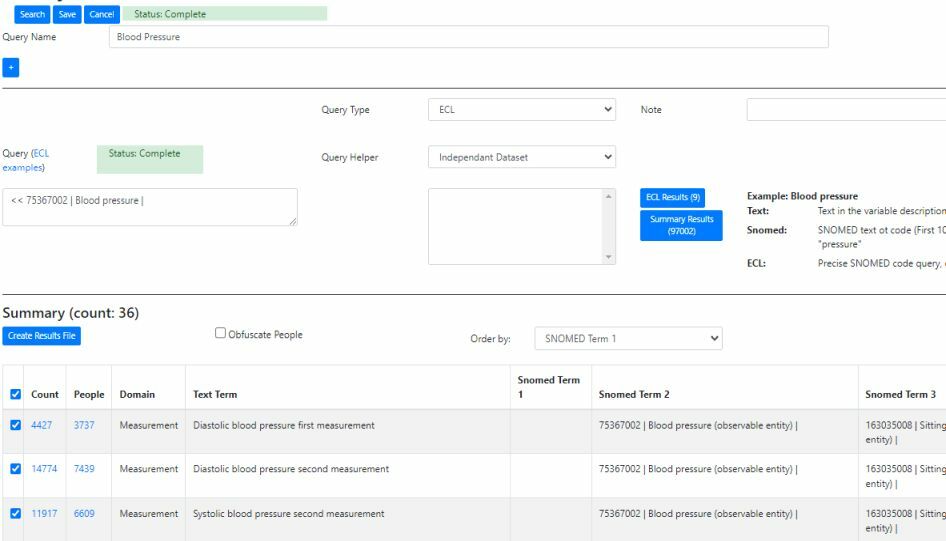
Query using NHS FHIR SNOMED CT API
The system supports the loading, storing, query and extraction of phenotype data. As Phenotypes vary greatly from measurements to questions, the Observational Medical Outcomes Partnership (OMOP) Common Data Model was used as an initial database design and then extended where necessary.
An Extract Transform & Load (ETL) solution maps incoming CSV format text files to the model. During the import process the phenotype data is indexed using both SNOMED CT terms and text.
The data can then be queried using a wildcard text query, SNOMED Expression Constraint Language (ECL) query and a hybrid “Snomed” approach comprising a wildcard query of any matching SNOMED CT terms followed by an ECL expansion query. SNOMED CT is a polyhierarchical thesaurus, where terms can have multiple parents and children. An ECL expansion query can be used to obtain results for any child terms (e.g. ECL query for “33468001 | Hematology procedure |” will pick up child terms such as “43396009 | Hemoglobin A1c measurement |”). This enables phenotype data to be indexed with low level / detailed terms and then retrieved using high level general terms.
SNOMED and ECL queries are via the NHS FHIR API (fast and currently free!)
Browse Phenotypes
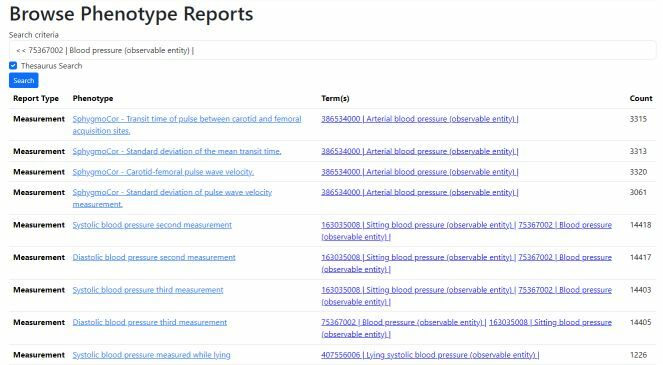
Search for dataset using text, SNOMED text or ECL query
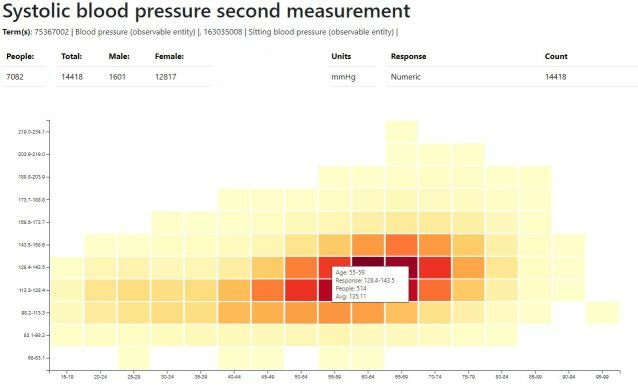
Example Report - Systolic Blood Pressure
A graphical reporting solution has been developed to display OMOP measurements and survey responses contained within the Phenobase repository. Over 4000 different sets of data can be located using text query or SNOMED ECL query and then displayed in 3 different graphs showing the distribution of data by age, the mean by age and discordance between twins. The solution currently works with 39 million measurements and survey responses, this is expected to double in the near term. See Phenobase Reporting White Paper for more details
Clinical Trial Registration
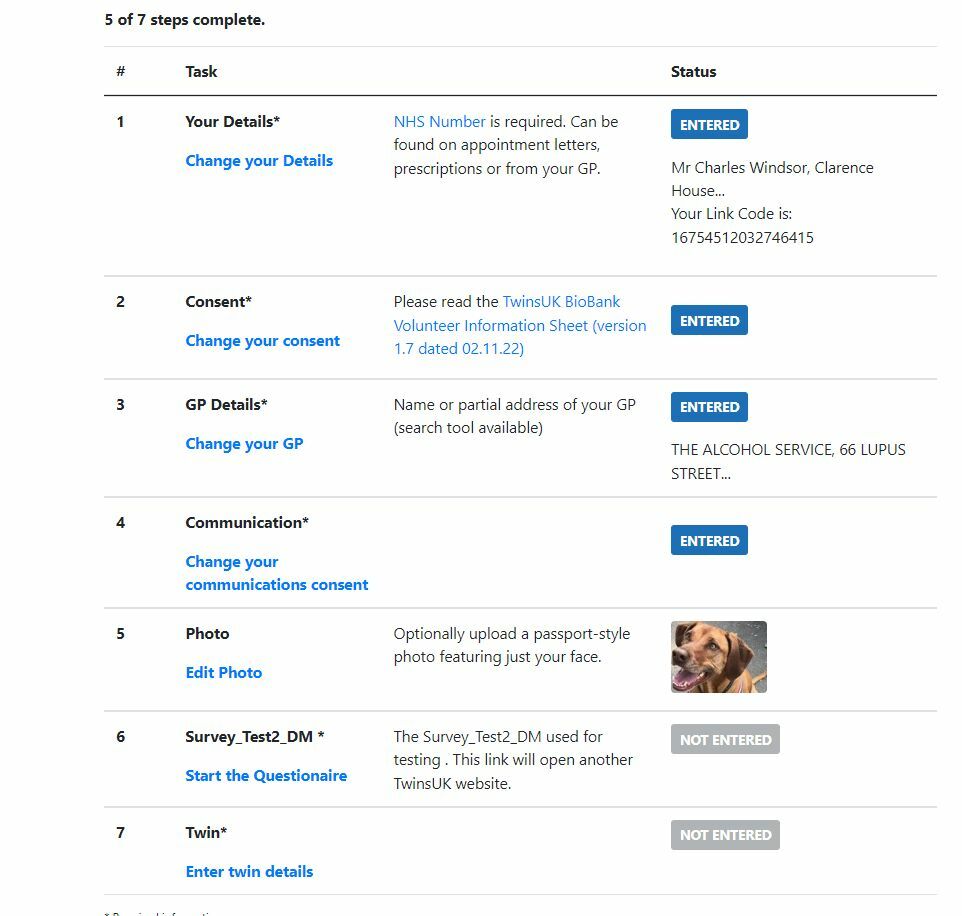
Medical Trial Registration
Registering for a clinical trial involves ensuring consent, gathering information, validating where possible (e.g. GP practice, postcodes, telephone numbers, emails, NHS numbers). This example system also includes automatic email reminders and matching of twins.
Data security is paramount in designing, building and running this system.
Clinic staff have access to an Admin view of the data enabling search, edit, matching twins manually etc.
The fully completed data can be extracted for the main department database.
The look and feel of the site is in line with other government sites.
Data Requests
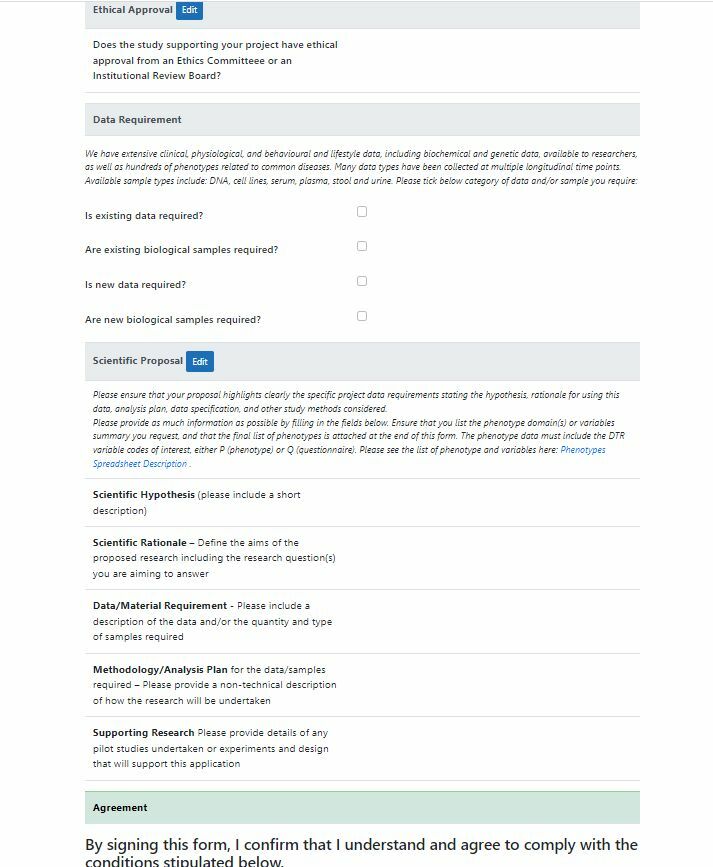
Phenotype Requests
Once data, samples (phenotypes) have been collected and collated, researchers can request access. This is a non-trivial process of collecting request details and authorisation.
Clinic staff have access to an Admin view of the data enabling search, edit and monitoring the process.
| Other Sections In This Site: |
|---|
| Sectors |
| Sectors / Clinical Data |
| Sectors / Bioinformatics |
| Sectors / Recruitment |
| Sectors / Billing |
| News |